Variables, Data Types, Indices, and Slices#
Assigning variables and using print() to check how the code is working#
To save a value, we assign them to a variable for later use. The syntax for assigning variables is: variable_name = variable_value.
Use print(variable_name) to print the specified variable.
Tips:
Choose informative names for variables.
Use comment lines to express the units of the variable or to describe the meaning of the variable.
Common operators are
+for addition,-for substraction,*for multiplication,/for division, and**for power operations.
Let’s see this in action with calculations using variables.
Exercise 1
How many grams of solid NaOH (40.0 g/mol) are required to prepare 500 ml of a 0.04 M solution?
Solution to Exercise 1
Here’s one possible solution.
liters = 0.5 #l
M = 0.04 #mol/l
MW = 40.0 #g/mol
wt = (liters * M) * MW #(l * mol/l) * g/mol = g
print(wt) #print the value that we calculated
Exercise 2
An enzyme has a \(V_{max}\) of 1.2 \(\mu\)\(M s^{-1}\) and a \(K_m\) of 10 \(\mu\)\(M\). What is the initial velocity (in \(\mu\)\(M s^{-1}\)) for an 8 \(\mu\)\(M\) substrate concentration?
Solution to Exercise 2
Here’s one possible solution.
Km = 10 #microM
Vmax = 1.2 #microM/s
S = 8 #microM
V0 = (Vmax * S) / (Km + S) #(microM/s * microM) / (microM + microM) = microM/s; Michaelis-Menten equation
print(V0) #print the value that we calculated
Exercise 3
Convert the initial velocity in \(\mu\)\(M s^{-1}\) from the previous exercise to \(\mu\)\(M min^{-1}\).
Solution to Exercise 3
Here’s one possible solution.
V0permin = V0 * 60 #microM/s * 60s/min = microM/min
print(V0permin) #print the value that we calculated
Data types, using len() to return the number of items in a sequence, and using .count() to count how many times an item appears in a sequence#
In Python, the data type is set when we assign a value to a variable. Different data types can do different things.
The most common data types are
strings (“str”) for text (surrounded by either single quotation marks or double quotation marks),
integers (“int”) for whole numbers, positive or negative, without decimals, of unlimited length,
floating point numbers (“float”) for numbers, positive or negative, containing one or more decimals,
lists (“list”) for multiple ordered and changeable items of different data types within one variable (created using square brackets (
[])),tuples (“tuple”) for multiple ordered and unchangeable items of different data types within one variable (created using round brackets (
())),dictionaries (“dict”) for storing a collection of ordered, changeable, and non-duplicate data as “key : value” pairs (created using curly brackets (
{}); pairs are separated using commas, and keys and values are separated using colons)
Use type(variable_name) to identify the data type of any variable.
Use len(variable_name) to determine the length of a sequence (e.g. a string, list, or tuple).
Use sequence_name.count(value) to count the number of items with a specified value within a sequence.
Use dictionary_name.get(key_name) to return the value of a specified key in a dictionary. If the key is not found, it returns None.
Let’s see this in action with DNA, RNA, and protein sequences as strings and lists with substrate concentrations and amino acids.
Exercise 4
Determine the data type, length, and number of tryptophan residues for this LRRK2 protein sequence containing one letter code amino acids.
protseqLRRK2 ="MASGSCQGCEEDEETLKKLIVRLNNVQEGKQIETLVQILEDLLVFTYSERASKLFQGKNIHVPLLIVLDSYMRVASVQQVGWSLLCKLIEVCPGTMQSLMGPQDVGNDWEVLGVHQLILKMLTVHNASVNLSVIGLKTLDLLLTSGKITLLILDEESDIFMLIFDAMHSFPANDEVQKLGCKALHVLFERVSEEQLTEFVENKDYMILLSALTNFKDEEEIVLHVLHCLHSLAIPCNNVEVLMSGNVRCYNIVVEAMKAFPMSERIQEVSCCLLHRLTLGNFFNILVLNEVHEFVVKAVQQYPENAALQISALSCLALLTETIFLNQDLEEKNENQENDDEGEEDKLFWLEACYKALTWHRKNKHVQEAACWALNNLLMYQNSLHEKIGDEDGHFPAHREVMLSMLMHSSSKEVFQASANALSTLLEQNVNFRKILLSKGIHLNVLELMQKHIHSPEVAESGCKMLNHLFEGSNTSLDIMAAVVPKILTVMKRHETSLPVQLEALRAILHFIVPGMPEESREDTEFHHKLNMVKKQCFKNDIHKLVLAALNRFIGNPGIQKCGLKVISSIVHFPDALEMLSLEGAMDSVLHTLQMYPDDQEIQCLGLSLIGYLITKKNVFIGTGHLLAKILVSSLYRFKDVAEIQTKGFQTILAILKLSASFSKLLVHHSFDLVIFHQMSSNIMEQKDQQFLNLCCKCFAKVAMDDYLKNVMLERACDQNNSIMVECLLLLGADANQAKEGSSLICQVCEKESSPKLVELLLNSGSREQDVRKALTISIGKGDSQIISLLLRRLALDVANNSICLGGFCIGKVEPSWLGPLFPDKTSNLRKQTNIASTLARMVIRYQMKSAVEEGTASGSDGNFSEDVLSKFDEWTFIPDSSMDSVFAQSDDLDSEGSEGSFLVKKKSNSISVGEFYRDAVLQRCSPNLQRHSNSLGPIFDHEDLLKRKRKILSSDDSLRSSKLQSHMRHSDSISSLASEREYITSLDLSANELRDIDALSQKCCISVHLEHLEKLELHQNALTSFPQQLCETLKSLTHLDLHSNKFTSFPSYLLKMSCIANLDVSRNDIGPSVVLDPTVKCPTLKQFNLSYNQLSFVPENLTDVVEKLEQLILEGNKISGICSPLRLKELKILNLSKNHISSLSENFLEACPKVESFSARMNFLAAMPFLPPSMTILKLSQNKFSCIPEAILNLPHLRSLDMSSNDIQYLPGPAHWKSLNLRELLFSHNQISILDLSEKAYLWSRVEKLHLSHNKLKEIPPEIGCLENLTSLDVSYNLELRSFPNEMGKLSKIWDLPLDELHLNFDFKHIGCKAKDIIRFLQQRLKKAVPYNRMKLMIVGNTGSGKTTLLQQLMKTKKSDLGMQSATVGIDVKDWPIQIRDKRKRDLVLNVWDFAGREEFYSTHPHFMTQRALYLAVYDLSKGQAEVDAMKPWLFNIKARASSSPVILVGTHLDVSDEKQRKACMSKITKELLNKRGFPAIRDYHFVNATEESDALAKLRKTIINESLNFKIRDQLVVGQLIPDCYVELEKIILSERKNVPIEFPVIDRKRLLQLVRENQLQLDENELPHAVHFLNESGVLLHFQDPALQLSDLYFVEPKWLCKIMAQILTVKVEGCPKHPKGIISRRDVEKFLSKKRKFPKNYMSQYFKLLEKFQIALPIGEEYLLVPSSLSDHRPVIELPHCENSEIIIRLYEMPYFPMGFWSRLINRLLEISPYMLSGRERALRPNRMYWRQGIYLNWSPEAYCLVGSEVLDNHPESFLKITVPSCRKGCILLGQVVDHIDSLMEEWFPGLLEIDICGEGETLLKKWALYSFNDGEEHQKILLDDLMKKAEEGDLLVNPDQPRLTIPISQIAPDLILADLPRNIMLNNDELEFEQAPEFLLGDGSFGSVYRAAYEGEEVAVKIFNKHTSLRLLRQELVVLCHLHHPSLISLLAAGIRPRMLVMELASKGSLDRLLQQDKASLTRTLQHRIALHVADGLRYLHSAMIIYRDLKPHNVLLFTLYPNAAIIAKIADYGIAQYCCRMGIKTSEGTPGFRAPEVARGNVIYNQQADVYSFGLLLYDILTTGGRIVEGLKFPNEFDELEIQGKLPDPVKEYGCAPWPMVEKLIKQCLKENPQERPTSAQVFDILNSAELVCLTRRILLPKNVIVECMVATHHNSRNASIWLGCGHTDRGQLSFLDLNTEGYTSEEVADSRILCLALVHLPVEKESWIVSGTQSGTLLVINTEDGKKRHTLEKMTDSVTCLYCNSFSKQSKQKNFLLVGTADGKLAIFEDKTVKLKGAAPLKILNIGNVSTPLMCLSESTNSTERNVMWGGCGTKIFSFSNDFTIQKLIETRTSQLFSYAAFSDSNIITVVVDTALYIAKQNSPVVEVWDKKTEKLCGLIDCVHFLREVMVKENKESKHKMSYSGRVKTLCLQKNTALWIGTGGGHILLLDLSTRRLIRVIYNFCNSVRVMMTAQLGSLKNVMLVLGYNRKNTEGTQKQKEIQSCLTVWDINLPHEVQNLEKHIEVRKELAEKMRRTSVE"
Solution to Exercise 4
Here’s one possible solution.
print(type(protseqLRRK2)) #determine and print the data type
len_protseqLRRK2 = len(protseqLRRK2) #determine the length of the string
print(len_protseqLRRK2) #print the value that we calculated
Wcount_protseqLRRK2 = protseqLRRK2.count("W") #count the number of times W appears in the string
print(Wcount_protseqLRRK2) #print the value that we calculated
Exercise 5
Define the EcoRI DNA recognition sequence (GAATTC) as a string.
Solution to Exercise 5
Here’s one possible solution.
DNAseqEcoRI = "GAATTC" #create a string using double quotation marks
Exercise 6
Determine the data type and length for this list with substrate concentrations.
subconc = [0, 1, 2, 4, 8, 15, 30, 60, 125, 250, 500]
Solution to Exercise 6
Here’s one possible solution.
print(type(subconc)) #determine and print the data type
len_subconc = len(subconc) #determine the length of the list
print(len_subconc) #print the value that we calculated
Exercise 7
Determine the data type, length, and number of alanine residues for this list with amino acids.
AA3Letter = ["ALA", "ARG", "ASN", "ASP", "CYS", "GLN", "GLU", "GLY", "HIS", "ILE", "LEU", "LYS", "MET", "PHE", "PRO", "SER", "THR", "TRP", "TYR", "VAL"]
Solution to Exercise 7
Here’s one possible solution.
print(type(AA3Letter)) #determine and print the data type
len_AA3Letter = len(AA3Letter) #determine the length of the list
print(len_AA3Letter) #print the value that we calculated
ALAcount_AA3Letter = AA3Letter.count("ALA") #count the number of times ALA appears in the string
print(ALAcount_AA3Letter) #print the value that we calculated
Exercise 8
Determine the number of “key : value” pairs in the following dictionary of restriction enzymes, with key = name and value = cleavage site. Retrieve the cleavage site for TaqI.
REs = {
'EcoRI' : 'GAATTC',
'BamHI' : 'GGATCC',
'EarI' : 'CTCTTC',
'ScaI' : 'AGTACT',
'NotI' : 'GCGGCCGC',
'TaqI' : 'TCGA',
'FokI' : 'GGATG',
'HindIII' : 'AAGCTT'
} #Create a dictionary with restriction enzyme names (keys) and cleavage sites (values). Both are strings.
Solution to Exercise 8
Here’s one possible solution.
len_REs = len(REs) #determine the length of the dictionary
print(len_REs) #print the value that we calculated
REs_TaqI = REs.get('TaqI') #look up the cleavage site stored for TaqI
print(REs_TaqI) #print the value that we calculated
Index and slice#
Sequence-based data types (e.g. a string, list, or tuple) are indexed, with the first item having index “0”. To select the first item, use sequence_name[0]; to select the second item, use sequence_name[1] …
If we have a long sequence and want to select an item towards the end, we can count backwards, starting at the index number “-1”.
The syntax for selecting a subset of an existing sequence, a slice, is: sequence_name[start:end]. When we specify the end item for the slice, it goes up to but does not include that item of the list!
Use sequence_name[:end] to have the slice starting from the beginning of the sequence.
Use sequence_name[start:] to have the slice going to the end of the sequence.
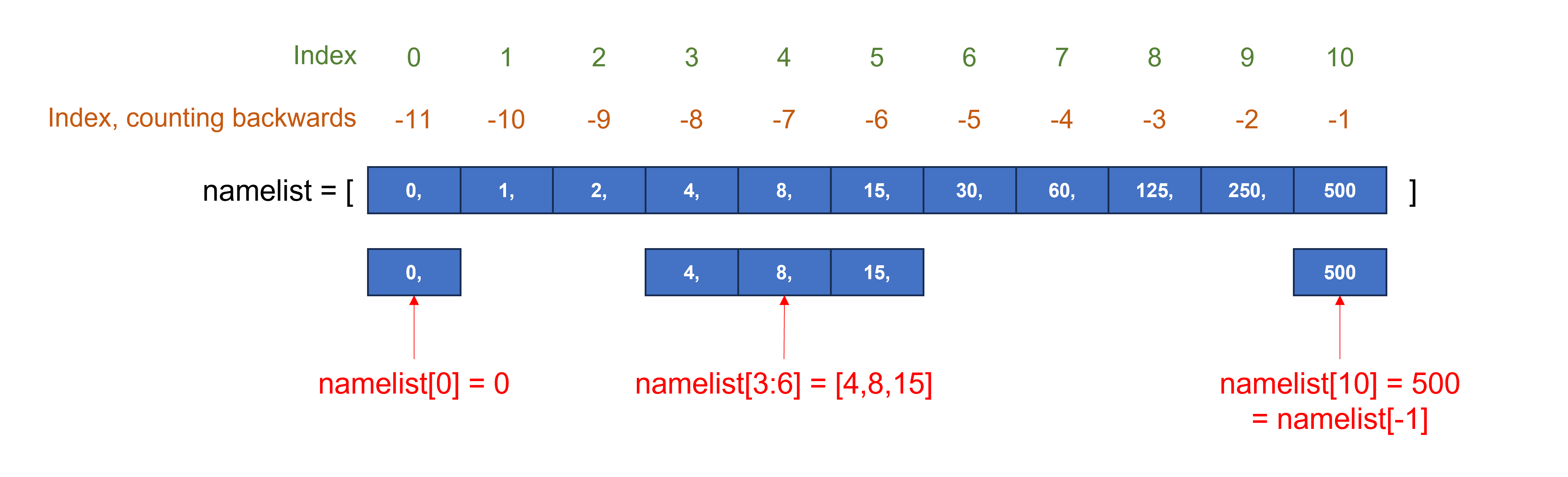
Let’s see this in action with DNA, RNA, and protein sequences as strings.
Exercise 9
Select the signal peptide (residues 1 to 22: MVSTMLSGLVLWLTFGWTPALA) and serine residues that are phosphorylated (S136 and S200) of this 7B2 protein sequence containing one letter code amino acids.
protseq7B2 ="MVSTMLSGLVLWLTFGWTPALAYSPRTPDRVSETDIQRLLHGVMEQLGIARPRVEYPAHQAMNLVGPQSIEGGAHEGLQHLGPFGNIPNIVAELTGDNTPKDFSEDQGYPDPPNPCPIGKTDDGCLENTPDTAEFSREFQLHQHLFDPEHDYPGLGKWNKKLLYEKMKGGQRRKRRSVNPYLQGQRLDNVVAKKSVPHFSDEDKDPE"
Solution to Exercise 9
Here’s one possible solution.
SPseq7B2 = protseq7B2[0:22] #Select residues 1 (0 as the first item has index 0) to 22 (22 as it goes up to but does not include item 22)
print(SPseq7B2) #print the value that we calculated
PhosS136 = protseq7B2[135] #Select residue 136 (135 as the first item has index 0)
print(PhosS136) #print the value that we calculated
PhosS200 = protseq7B2[199] #Select residue 200 (199 as the first item has index 0)
print(PhosS200) #print the value that we calculated