Data Types, Index, and Slice#
Data types#
In Python, the data type is set when we assign a value to a variable. Different data types can do different things.
The most common data types are
Strings (
str) for text (surrounded by either single quotation marks or double quotation marks)Integers (
int) for whole numbers, positive or negative, without decimals, of unlimited lengthFloating point numbers (
float) for numbers, positive or negative, containing one or more decimalsBooleans (
bool), with only two possible values:trueandfalseLists (
list) for multiple ordered and changeable items of different data types within one variable (created using square brackets)Tuples (
tuple) for multiple ordered and unchangeable items of different data types within one variable (created using round brackets)Dictionaries (
dict) for storing a collection of ordered, changeable, and non-duplicate data as “key : value” pairs (created using curly brackets; pairs are separated using commas, and keys and values are separated using colons)
Use type(variable_name) to identify the data type of any variable.
Use len(sequence_name) to determine the length of a sequence (e.g. a string, list, or tuple). Use sequence_name.count(value) to count the number of items with a specified value within a sequence.
Use sorted(list_name, key=myFunc (optional), reverse=True (optional)) to sort a list in ascending (by default), descending (optional) or user-defined (optional) order. The original list variable is unchanged. Of note, strings are sorted in alphabetical order based on their first letter (A-Z). However, words that start with uppercase letters come before words that start with lowercase letters. For case-insensitive sorting, set the key argument to key=str.lower, which converts all strings to lowercase characters.
Use dictionary_name.get(key_name) to return the value of a specified key in a dictionary. If the key is not found, it returns None.
Index and slice for sequence-based data types#
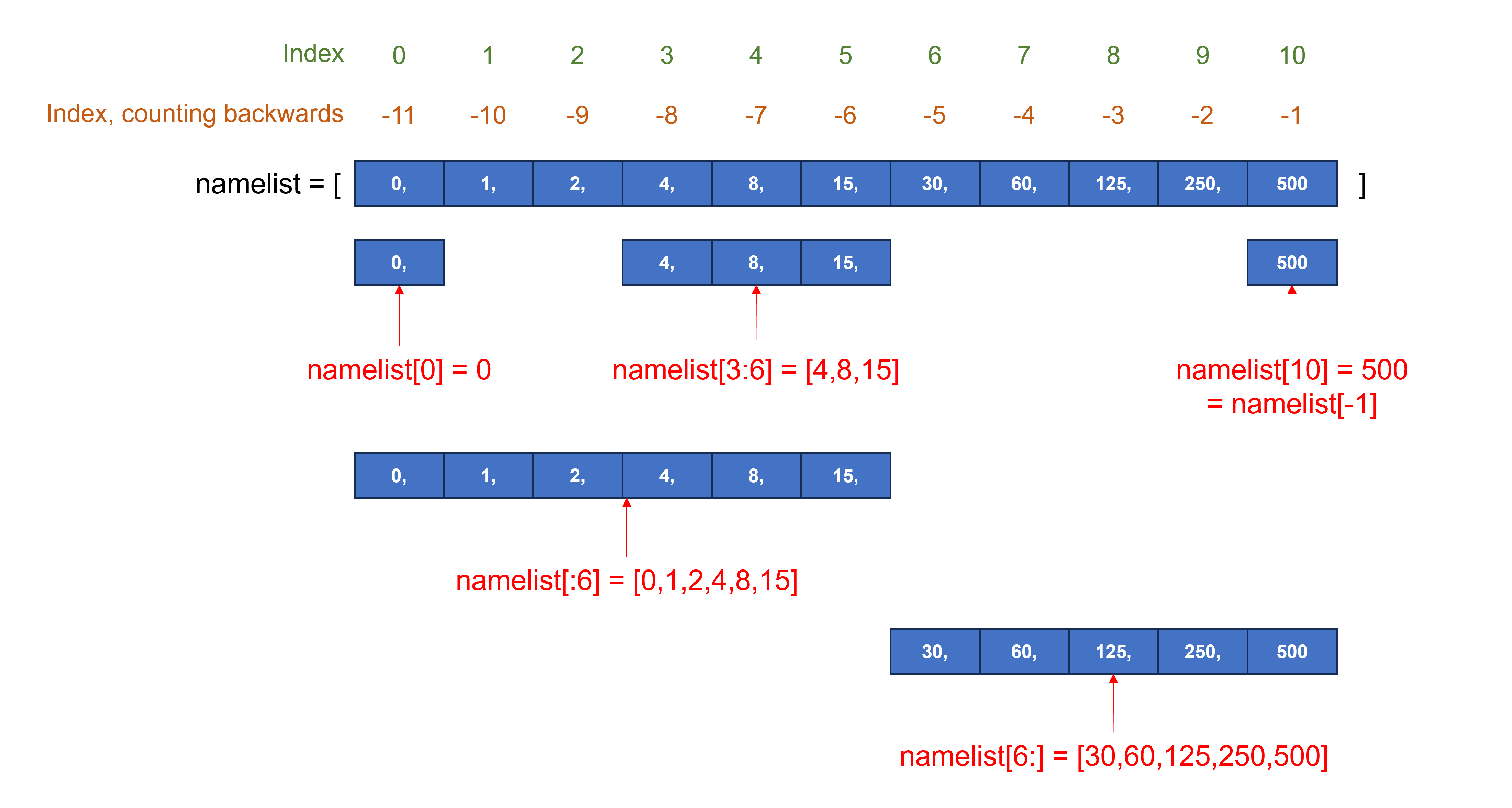
Sequence-based data types (e.g. a string, list, or tuple) are indexed, with _the first item having index 0. To select the first item, use sequence_name[0]; to select the second item, use sequence_name[1] …
If we have a long sequence and want to select an item towards the end, we can count backwards, starting at the index number -1.
The syntax for selecting a subset of an existing sequence, a slice, is: sequence_name[start:end]. When we specify the end item for the slice, it goes up to but does not include that item of the sequence!
Use sequence_name[:end] to have the slice starting from the beginning of the sequence. Use sequence_name[start:] to have the slice going to the end of the sequence.
The syntax for selecting a slice with regular steps, is: sequence_name[start:end:step]. A negative step goes backwards. For example, use sequence_name[::2] to select every other element from the sequence, and use sequence_name[::-1] to select the elements in reverse order.
Examples#
Please pay attention to the use of comments (with #) to express the units of variables or to describe the meaning of commands.
Example
Determine the data type, length, and number of tryptophan residues for this LRRK2 protein sequence containing one letter code amino acids. Select residue 2019. The G2019S mutation in LRRK2 is the most common genetic determinant of Parkinson’s disease identified to date. It lies in the protein’s kinase domain. Select this kinase domain, it includes residue 1879 to residue 2138.
protseqLRRK2 = "MASGSCQGCEEDEETLKKLIVRLNNVQEGKQIETLVQILEDLLVFTYSERASKLFQGKNIHVPLLIVLDSYMRVASVQQVGWSLLCKLIEVCPGTMQSLMGPQDVGNDWEVLGVHQLILKMLTVHNASVNLSVIGLKTLDLLLTSGKITLLILDEESDIFMLIFDAMHSFPANDEVQKLGCKALHVLFERVSEEQLTEFVENKDYMILLSALTNFKDEEEIVLHVLHCLHSLAIPCNNVEVLMSGNVRCYNIVVEAMKAFPMSERIQEVSCCLLHRLTLGNFFNILVLNEVHEFVVKAVQQYPENAALQISALSCLALLTETIFLNQDLEEKNENQENDDEGEEDKLFWLEACYKALTWHRKNKHVQEAACWALNNLLMYQNSLHEKIGDEDGHFPAHREVMLSMLMHSSSKEVFQASANALSTLLEQNVNFRKILLSKGIHLNVLELMQKHIHSPEVAESGCKMLNHLFEGSNTSLDIMAAVVPKILTVMKRHETSLPVQLEALRAILHFIVPGMPEESREDTEFHHKLNMVKKQCFKNDIHKLVLAALNRFIGNPGIQKCGLKVISSIVHFPDALEMLSLEGAMDSVLHTLQMYPDDQEIQCLGLSLIGYLITKKNVFIGTGHLLAKILVSSLYRFKDVAEIQTKGFQTILAILKLSASFSKLLVHHSFDLVIFHQMSSNIMEQKDQQFLNLCCKCFAKVAMDDYLKNVMLERACDQNNSIMVECLLLLGADANQAKEGSSLICQVCEKESSPKLVELLLNSGSREQDVRKALTISIGKGDSQIISLLLRRLALDVANNSICLGGFCIGKVEPSWLGPLFPDKTSNLRKQTNIASTLARMVIRYQMKSAVEEGTASGSDGNFSEDVLSKFDEWTFIPDSSMDSVFAQSDDLDSEGSEGSFLVKKKSNSISVGEFYRDAVLQRCSPNLQRHSNSLGPIFDHEDLLKRKRKILSSDDSLRSSKLQSHMRHSDSISSLASEREYITSLDLSANELRDIDALSQKCCISVHLEHLEKLELHQNALTSFPQQLCETLKSLTHLDLHSNKFTSFPSYLLKMSCIANLDVSRNDIGPSVVLDPTVKCPTLKQFNLSYNQLSFVPENLTDVVEKLEQLILEGNKISGICSPLRLKELKILNLSKNHISSLSENFLEACPKVESFSARMNFLAAMPFLPPSMTILKLSQNKFSCIPEAILNLPHLRSLDMSSNDIQYLPGPAHWKSLNLRELLFSHNQISILDLSEKAYLWSRVEKLHLSHNKLKEIPPEIGCLENLTSLDVSYNLELRSFPNEMGKLSKIWDLPLDELHLNFDFKHIGCKAKDIIRFLQQRLKKAVPYNRMKLMIVGNTGSGKTTLLQQLMKTKKSDLGMQSATVGIDVKDWPIQIRDKRKRDLVLNVWDFAGREEFYSTHPHFMTQRALYLAVYDLSKGQAEVDAMKPWLFNIKARASSSPVILVGTHLDVSDEKQRKACMSKITKELLNKRGFPAIRDYHFVNATEESDALAKLRKTIINESLNFKIRDQLVVGQLIPDCYVELEKIILSERKNVPIEFPVIDRKRLLQLVRENQLQLDENELPHAVHFLNESGVLLHFQDPALQLSDLYFVEPKWLCKIMAQILTVKVEGCPKHPKGIISRRDVEKFLSKKRKFPKNYMSQYFKLLEKFQIALPIGEEYLLVPSSLSDHRPVIELPHCENSEIIIRLYEMPYFPMGFWSRLINRLLEISPYMLSGRERALRPNRMYWRQGIYLNWSPEAYCLVGSEVLDNHPESFLKITVPSCRKGCILLGQVVDHIDSLMEEWFPGLLEIDICGEGETLLKKWALYSFNDGEEHQKILLDDLMKKAEEGDLLVNPDQPRLTIPISQIAPDLILADLPRNIMLNNDELEFEQAPEFLLGDGSFGSVYRAAYEGEEVAVKIFNKHTSLRLLRQELVVLCHLHHPSLISLLAAGIRPRMLVMELASKGSLDRLLQQDKASLTRTLQHRIALHVADGLRYLHSAMIIYRDLKPHNVLLFTLYPNAAIIAKIADYGIAQYCCRMGIKTSEGTPGFRAPEVARGNVIYNQQADVYSFGLLLYDILTTGGRIVEGLKFPNEFDELEIQGKLPDPVKEYGCAPWPMVEKLIKQCLKENPQERPTSAQVFDILNSAELVCLTRRILLPKNVIVECMVATHHNSRNASIWLGCGHTDRGQLSFLDLNTEGYTSEEVADSRILCLALVHLPVEKESWIVSGTQSGTLLVINTEDGKKRHTLEKMTDSVTCLYCNSFSKQSKQKNFLLVGTADGKLAIFEDKTVKLKGAAPLKILNIGNVSTPLMCLSESTNSTERNVMWGGCGTKIFSFSNDFTIQKLIETRTSQLFSYAAFSDSNIITVVVDTALYIAKQNSPVVEVWDKKTEKLCGLIDCVHFLREVMVKENKESKHKMSYSGRVKTLCLQKNTALWIGTGGGHILLLDLSTRRLIRVIYNFCNSVRVMMTAQLGSLKNVMLVLGYNRKNTEGTQKQKEIQSCLTVWDINLPHEVQNLEKHIEVRKELAEKMRRTSVE" #create a string using double quotation marks
type_protseqLRRK2 = type(protseqLRRK2) #determine the data type
len_protseqLRRK2 = len(protseqLRRK2) #determine the length of the string
Wcount_protseqLRRK2 = protseqLRRK2.count("W") #count the number of times W appears in the string
res2019_protseqLRRK2 = protseqLRRK2[2018] #select residue 2019 (2018 as the first item has index 0)
kinase_protseqLRRK2 = protseqLRRK2[1878:2138] #select residues 1879 (1878 as the first item has index 0) to 2138 (the first item has index 0; however, it goes up to but does not include item 2138)
print("This variable has data type", type_protseqLRRK2, ".",
"The length of the sequence is", len_protseqLRRK2, "residues .",
"Trp appears", Wcount_protseqLRRK2, "times .",
"Amino acid 2019 is a", res2019_protseqLRRK2, ".",
"The sequence of the LRRK2 kinase domain is", kinase_protseqLRRK2, ".") #Print and place all values that we calculated in a readable sentence. The print function can take more than one object. By default, it separates objects with a space.
This variable has data type <class 'str'> . The length of the sequence is 2527 residues . Trp appears 26 times . Amino acid 2019 is a G . The sequence of the LRRK2 kinase domain is QAPEFLLGDGSFGSVYRAAYEGEEVAVKIFNKHTSLRLLRQELVVLCHLHHPSLISLLAAGIRPRMLVMELASKGSLDRLLQQDKASLTRTLQHRIALHVADGLRYLHSAMIIYRDLKPHNVLLFTLYPNAAIIAKIADYGIAQYCCRMGIKTSEGTPGFRAPEVARGNVIYNQQADVYSFGLLLYDILTTGGRIVEGLKFPNEFDELEIQGKLPDPVKEYGCAPWPMVEKLIKQCLKENPQERPTSAQVFDILNSAELV .
Example
Determine the number of “key : value” pairs in the following dictionary with genetic codes, with key = code and value = amino acid. Retrieve the genetic code translation for TTC.
GeneticCode = {
'ATA':'I', 'ATC':'I', 'ATT':'I', 'ATG':'M',
'ACA':'T', 'ACC':'T', 'ACG':'T', 'ACT':'T',
'AAC':'N', 'AAT':'N', 'AAA':'K', 'AAG':'K',
'AGC':'S', 'AGT':'S', 'AGA':'R', 'AGG':'R',
'CTA':'L', 'CTC':'L', 'CTG':'L', 'CTT':'L',
'CCA':'P', 'CCC':'P', 'CCG':'P', 'CCT':'P',
'CAC':'H', 'CAT':'H', 'CAA':'Q', 'CAG':'Q',
'CGA':'R', 'CGC':'R', 'CGG':'R', 'CGT':'R',
'GTA':'V', 'GTC':'V', 'GTG':'V', 'GTT':'V',
'GCA':'A', 'GCC':'A', 'GCG':'A', 'GCT':'A',
'GAC':'D', 'GAT':'D', 'GAA':'E', 'GAG':'E',
'GGA':'G', 'GGC':'G', 'GGG':'G', 'GGT':'G',
'TCA':'S', 'TCC':'S', 'TCG':'S', 'TCT':'S',
'TTC':'F', 'TTT':'F', 'TTA':'L', 'TTG':'L',
'TAC':'Y', 'TAT':'Y', 'TAA':'_', 'TAG':'_',
'TGC':'C', 'TGT':'C', 'TGA':'_', 'TGG':'W',
}
len_GeneticCode = len(GeneticCode) #determine the length of the dictionary
GeneticCode_TTC = GeneticCode.get('TTC') #look up the cleavage site stored for NotI
print("This dictionary contains", len_GeneticCode, "pairs .",
"The genetic code translation stored for TTC is", GeneticCode_TTC, ".") #Print and place all values that we calculated in a readable sentence. The print function can take more than one object. By default, it separates objects with a space.
This dictionary contains 64 pairs . The genetic code translation stored for TTC is F .
Example
Determine the data type and length for this list with amino acids. Sort the list in alphabetical order.
AA3Letter = ["ALA", "ARG", "ASN", "ASP", "CYS", "GLN", "GLU", "GLY", "HIS", "ILE", "LEU", "LYS", "MET", "PHE", "PRO", "SER", "THR", "TRP", "TYR", "VAL"]
type_AA3Letter = type(AA3Letter) #determine and print the data type
len_AA3Letter = len(AA3Letter) #determine the length of the list
AA3Lettersorted = sorted(AA3Letter, key=str.lower) #sort the list in ascending order
print("This variable has data type", type_AA3Letter, ".",
"This list contains", len_AA3Letter, "amino acids .",
"After sorting the concentrations in ascending order, this list becomes", AA3Lettersorted, ".") #Print and place all values that we calculated in a readable sentence. The print function can take more than one object. By default, it separates objects with a space.
This variable has data type <class 'list'> . This list contains 20 amino acids . After sorting the concentrations in ascending order, this list becomes ['ALA', 'ARG', 'ASN', 'ASP', 'CYS', 'GLN', 'GLU', 'GLY', 'HIS', 'ILE', 'LEU', 'LYS', 'MET', 'PHE', 'PRO', 'SER', 'THR', 'TRP', 'TYR', 'VAL'] .
Exercises#
Exercise
Define the EcoRI DNA recognition sequence (GAATTC) as a string. Determine length and number of adenine bases for this sequence.
Solution
Here’s one possible solution.
DNAseqEcoRI = "GAATTC" #create a string using double quotation marks
len_DNAseqEcoRI = len(DNAseqEcoRI) #determine the length of the string
Acount_DNAseqEcoRI = DNAseqEcoRI.count("A") #count the number of times A appears in the string
print("The length of the sequence is", len_DNAseqEcoRI, "residues .",
"Adenosine appears", Acount_DNAseqEcoRI, "times .") #Print and place all values that we calculated in a readable sentence. The print function can take more than one object. By default, it separates objects with a space.
Exercise
Calculate the melting temperature of the primer with sequence “GACTGCGTTAGGATTGGC”. The melting temperature (°C) can be computed as 64.9 + 41 x (GC - 16.4)/N, with GC the total number of G and C bases in the primer and N the primer length.
Solution
Here’s one possible solution.
primer = "GACTGCGTTAGGATTGGC" #create a string using double quotation marks
N_primer = len(primer) #determine the length of the string
G_primer = primer.count("G") #count the number of times G appears in the string
C_primer = primer.count("C") #count the number of times C appears in the string
GC_primer = G_primer + C_primer #calculate the number of times G and C appear in the string
Tm_primer = 64.9 + 41 * (GC_primer - 16.4)/N_primer #calculate the melting temperature using the formula
print("The melting temperature for primer", primer, "is", Tm_primer, "°C .") #Print and place all values that we calculated in a readable sentence. The print function can take more than one object. By default, it separates objects with a space.
Exercise
Determine the data type and length for this list with substrate concentrations. Sort the list in ascending order.
subconc = [1, 4, 500, 0, 15, 250, 30, 125, 2, 8, 60]
Solution
Here’s one possible solution.
type_subconc = type(subconc) #determine and print the data type
len_subconc = len(subconc) #determine the length of the list
subconcsorted = sorted(subconc) #sort the list in ascending order
print("This variable has data type", type_subconc, ".",
"This list contains", len_subconc, "concentrations .",
"After sorting the concentrations in ascending order, this list becomes", subconcsorted, ".") #Print and place all values that we calculated in a readable sentence. The print function can take more than one object. By default, it separates objects with a space.
Exercise
Determine the number of “key : value” pairs in the following dictionary of restriction enzymes, with key = name and value = cleavage site. Retrieve the cleavage site for NotI.
REs = {
'EcoRI' : 'GAATTC',
'BamHI' : 'GGATCC',
'EarI' : 'CTCTTC',
'ScaI' : 'AGTACT',
'NotI' : 'GCGGCCGC',
'TaqI' : 'TCGA',
'FokI' : 'GGATG',
'HindIII' : 'AAGCTT'
} #Create a dictionary with restriction enzyme names (keys) and cleavage sites (values). Both are strings.
Solution
Here’s one possible solution.
len_REs = len(REs) #determine the length of the dictionary
REs_NotI = REs.get('NotI') #look up the cleavage site stored for NotI
print("This dictionary contains", len_REs, "pairs .",
"The cleavage site stored for NotI is", REs_NotI, ".") #Print and place all values that we calculated in a readable sentence. The print function can take more than one object. By default, it separates objects with a space.
